Introduction:
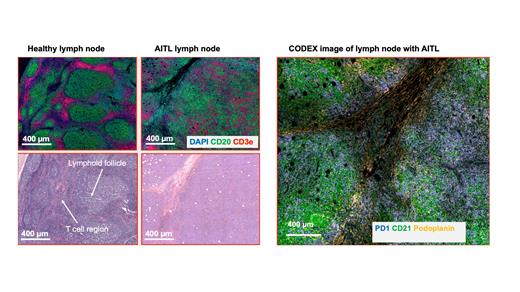
Angioimmunoblastic T-cell lymphoma (AITL) is a subtype of non-Hodgkin lymphoma (NHL) characterized primarily by a T follicular helper cell (TFH) phenotype. AITL presents with a highly complex tumor microenvironment with the tumor cells on a scaffold of highly arborized CD21+ follicular dendritic cells found in proximity to high endothelial venules (HEVs). The organized structure of the lymph node with distinct B cell and T cell zones is disrupted in the lymph node samples from patients with AITL. Lymphoid follicles in most AITL cases tend to be depleted, not hyperplastic as in other lymphomas. AITL tumor cells express CD10, BCL6, PD1, CXCL13, C-X-C motif chemokine receptor 5 (CXCR5), ICOS, and signaling lymphocytic activation molecule-associated protein (SAP) with diagnosis based on the expression of at least 2 of these markers. Our study uses novel spatial omics technology (Deterministic Barcoding in Tissue (DBiT-seq) and multiplexed immunofluorescence staining (CODEX)) to fully characterize the transcriptional and proteomic landscape of AITL.
Methods:
In this study, initial FFPE biopsy specimens from 22 patients with AITL were collected from archives. 10 µm thick tissue sections on poly-L-lysine coated slides were prepared for the performance of multiplexed immunofluorescence imaging (CODEX) for 12 immune-related markers: Podoplanin, CD20, CD4, CD8, CD68, CD45RO, CD3e, GranzymeB, PD1, Ki67, CD21 and CD34 using the PhenoCycler-Fusion system. We also validated anti-EBV Latent Membrane Protein 1 antibody. Cell segmentation was performed using a Stardist-based model in QuPath and downstream analysis done using the Seurat package. On an adjacent tissue section, we used Spatial CITE-seq to co-map the whole transcriptome and ~160 proteins.
Results:
From the multiplexed immunofluorescence staining, we observed CD21+ Follicular dendritic cells in close proximity to HEVs as expected from the spatial architecture of AITL. We also found 11 spatially distinct clusters differentially expressing the marker proteins. We observed a lower number of CD8+ cytotoxic T cells relative to CD4+ Helper T cells. We also identified cellular neighborhoods and their composition found a population of FDCs that directly interact with the tumor cells. From spatial CITE-seq, we average of ~670 genes were found in each spot. We also found the expression of CXCL13, TET2, DNMT3A and PDCP1 which are markers associated with AITL. There was a strong correlation between the transcript signals detected in Spatial CITE-seq and the protein expression from CODEX.
Conclusions:
Our study revealed the spatial organization of the tumor microenvironment of AITL. We envision that combining multiplexed immunofluorescence and Spatial CITE-seq will further our understanding of the AITL tumor environment and could lead to the discovery of novel clinical targets.
Disclosures
Fan:IsoPlexis: Membership on an entity's Board of Directors or advisory committees; Singleron Biotechnologies: Membership on an entity's Board of Directors or advisory committees; AtlasXomics: Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal